rsync, please read its excellent man page.Table of Contents
Goethe-NHR Cluster Usage
Goethe-NHR is a general-purpose computer cluster running AlmaLinux 9 and SLURM. Please read the following instructions and ensure that this guide is fully understood before using the system.
Login
An SSH client is required to connect to the cluster. On a Linux system the command is usually:
ssh <user_account>@goethe.hhlr-gu.de
Am I connected to the right server? Please find additional information here:
goethe.hhlr-gu.de fingerprints
You may receive a warning from your system that something with the security is wrong, “maybe somebody is evedropping”. Due to the upgrade of the operating system from Scientifc Linux 7.9 to AlmaLinux 9.2 the SSH server keys have changed. Please erase your old Goethe key with
ssh-keygen -R goethe.hhlr-gu.de
and accept the new goethe.hhlr-gu.de key. Above you will find our unique ECDSA and ED25519 fingerprints. Some programs tend to display the fingerprint in SHA256 or MD5 format. Just click on goethe.hhlr-gu.de fingerprints above.
On Windows systems please use/install a Windows SSH client (e.g. PuTTY, MobaXterm, the Cygwin ssh package or the built-in ssh command).
During your first login you will get the message, that your password has expired and you have to change it. Re-enter the initial password provided by the CSC, then choose a new one and confirm it. You will be logged out automatically. Now you can login with your new password and work on the cluster.
ulimit -t
on the command line. On a login node, any process that exceeds the CPU-time limit (e.g. a long running test program or a long running rsync) will be killed automatically.
Environment Modules
There are several versions of software packages installed on our systems. The same name for an executable (e.g. mpirun) and/or library file may be used by more than one package. The environment module system, with its module command, helps to keep them apart and prevents name clashes. You can list the module-managed software by running module avail on the command line. Other important commands are module load <name> (loads a module) and module list (lists the already loaded modules).
If you want to know more about module commands, the module help command will give you an overview.
Working with Intel oneAPI
With the command 'module avail' you are able to see all available modules on the cluster. The intel/oneapi/xxx works a kind of different, because it functions like a container. Start loading it with module load intel/oneapi/xxx and see with module avail what is inside. Now start loading your preferred module.
latest.
Example
module load intel/oneapi/2023.2.0 module avail ... # new modules available within the Intel oneAPI modulefile ----------------------------------- /cluster/intel/oneapi/2023.2.0/modulefiles --------------------- advisor/2023.2.0 dev-utilities/2021.10.0 icc32/2023.2.1 mkl32/2023.2.0 advisor/latest dev-utilities/latest icc32/latest mkl32/latest ccl/2021.10.0 dnnl-cpu-gomp/2023.2.0 inspector/2023.2.0 mpi/2021.10.0 ccl/latest dnnl-cpu-gomp/latest inspector/latest mpi/latest compiler-rt/2023.2.1 dnnl-cpu-iomp/2023.2.0 intel_ipp_ia32/2021.9.0 mpi_BROKEN/2021.10.0 compiler-rt/latest dnnl-cpu-iomp/latest intel_ipp_ia32/latest mpi_BROKEN/latest compiler-rt32/2023.2.1 dnnl-cpu-tbb/2023.2.0 intel_ipp_intel64/2021.9.0 oclfpga/2023.2.0 compiler-rt32/latest dnnl-cpu-tbb/latest intel_ipp_intel64/latest oclfpga/2023.2.1 compiler/2023.2.1 dnnl/2023.2.0 intel_ippcp_ia32/2021.8.0 oclfpga/latest compiler/latest dnnl/latest intel_ippcp_ia32/latest tbb/2021.10.0 compiler32/2023.2.1 dpct/2023.2.0 intel_ippcp_intel64/2021.8.0 tbb/latest compiler32/latest dpct/latest intel_ippcp_intel64/latest tbb32/2021.10.0 dal/2023.2.0 dpl/2022.2.0 itac/2021.10.0 tbb32/latest dal/latest dpl/latest itac/latest vtune/2023.2.0 debugger/2023.2.0 icc/2023.2.1 mkl/2023.2.0 vtune/latest debugger/latest icc/latest mkl/latest Key: loaded modulepath
Please also note, by default, Intel MPI's mpicc uses the GCC. To make Intel MPI use an Intel compiler you have to set I_MPI_CC in your environment (or use mpiicc), e.g.:
module load intel/oneapi/2023.2.0 module load compiler/2023.2.1 module load mpi/2021.10.0 export I_MPI_CC=icx
Compiling Software
You can compile your software on the login nodes (or on any other node, inside a job allocation). Several compiler suites are available. While GCC version 11.X.Y is the built-in OS default, you can list additional compilers and libraries by running module avail:
- Intel compilers
- MPI libraries
- other libraries
For the right compilation commands please consider:
To build and manage software which is not available via “module avail” and is not available as a built-in OS package, we recommend using Spack. Please read this small introduction on how to use Spack on the cluster. More information is available on the Spack webpage.
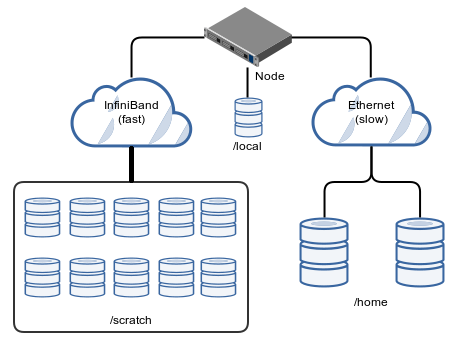
Storage
There are various storage systems available on the cluster. In this section we describe the most relevant:
- your home directory
/home/<group>/<user>(NFS, slow), - your scratch directory
/scratch/<group>/<user>(parallel file system BeeGFS, fast), - the non-shared local storage (i.e. unique on each compute node) under
/local/$SLURM_JOB_IDon each compute node (max. 1.4 TB per node, slow) - and the two archive file systems
/arc01and/arc02(explained at the end of this section).
Please use your home directory for small permanent files, e.g. source files, libraries and executables.
By default, the space in your home directory is limited to 30 GB, and in your scratch directory to 5 TB and/or 800000 inodes (which corresponds to approximately 200000+ files). You can check your homedir and scratch usage by running the quota command on a login node.
If you need local storage on the compute nodes, you have to add the --tmp parameter to your job script (see SLURM section below). Set the amount of storage in megabytes, e.g. set --tmp=5000 to allocate 5 GB of local disk space. The local directory (/local/$SLURM_JOB_ID) is deleted after the corresponding job has finished. If, for some reason, you don't want the data to be deleted (e.g. for debugging), you can use salloc instead of sbatch and work interactively (see man salloc). Or, one can put an rsync at the end of the job script, in order to save the local data to /scratch just before the job exits:
... mkdir /scratch/<groupid>/<userid>/$SLURM_JOBID scontrol show hostnames $SLURM_JOB_NODELIST | xargs -i ssh {} \ rsync -a /local/$SLURM_JOBID/ \ /scratch/<groupid>/<userid>/$SLURM_JOBID/{}
In addition to the “volatile” /scratch and the permanent /home, which come along with every user account, more permanent disk space (2 × N, where N ≤ 10 TB) can be requested by group leaders for archiving. Upon request, two file systems will be created for every group member, to be accessed through rsync1), e.g. list the contents of your folder and archive a /scratch directory:
rsync arc01:/archive/<group>/<user>/ ... cd /scratch/<group>/<user>/ rsync [--progress] -a <somefolder> arc01:/archive/<group>/<user>/
or, for arc02:
rsync arc02:/archive/<group>/<user>/ ... cd /scratch/<group>/<user>/ rsync [--progress] -a <somefolder> arc02:/archive/<group>/<user>/
The space is limited by N on each of the both systems. Limits are set for an entire group (there's no user quota). The disk usage can be checked by running
df -h /arc0{1,2}/archive/<group>
on the command line. The corresponding hardware resides in separate server rooms. There is no automatic backup. However, for a user, a possible backup scenario is to backup his or her data manually to both storage systems, arc01 and arc02 (e.g. at the end of a compute job). Note: The archive file systems are mounted on the login nodes, but not on the compute nodes. So it's not possible to use the archive for direct job I/O. Please use rsync as described above.
- All shared file systems are shared between users and jobs. There is no guarantee, that you always get the desired bandwidth and/or response time.
- Although our storage systems are protected by RAID mechanisms, we can't guarantee the safety of your data. It is within the responsibility of the user to backup important files.
Running Jobs With SLURM
On our systems, compute jobs and resources are managed by SLURM (Simple Linux Utility for Resource Management). The compute nodes are organized in partitions (or queues) named general1 and gpu. There is also a small test partition called test. Additionally we offer the use of some old compute nodes from the former LOEWE-CSC cluster. Those nodes are organized in the partition general2. You can see more details (the current number of nodes in each partition and their state) by running the sinfo command on a login node.
| Partition | CPU | GPU |
|---|---|---|
general1 | Intel Skylake CPU | n/a |
general2 | Intel Ivy Bridge CPU Intel Broadwell CPU | n/a |
gpu | AMD EPYC 7452 | AMD Instinct MI210 |
test | Intel Skylake CPU | n/a |
Nodes are used exclusively, i.e. only whole nodes are allocated for a job and no other job can use the same nodes concurrently.
In this document we discuss several job types and use cases. In most cases, a compute job falls under one (or more than one) of the following categories:
For every compute job you have to submit a job script (unless working interactively using salloc or srun, see man page for more information). If jobscript.sh is such a script, then a job can be enqueued by running
sbatch jobscript.sh
on a login node. A SLURM job script is a shell script containing SLURM directives (options), i.e. pseudo-comment lines starting with
#SBATCH ...
The SLURM options define the resources to be allocated for the job (and some other properties). Otherwise the script contains the “job logic”, i.e. commands to be executed.
Read More
The following instructions shall provide you with the basic information you need to get started with SLURM on our systems. However, the official SLURM documentation covers some more use cases (also in more detail). Please read the SLURM man pages (e.g. man sbatch or man salloc) and/or visit http://www.schedmd.com/slurmdocs. It's highly recommended.
The test Partition: Your First Job Script
You can use the (very small) test partition for pre-production or tests. In test you can run jobs with a walltime of no longer than two hours. In the following example we allocate 160 CPU tasks (i.e. 160 CPU threads on two nodes = 80 tasks per node) and 512 MB per task for 5 minutes (SLURM may kill the job after that time, if it's still running):
#!/bin/bash #SBATCH --job-name=foo #SBATCH --partition=test #SBATCH --nodes=2 #SBATCH --ntasks=160 #SBATCH --cpus-per-task=1 #SBATCH --mem-per-cpu=512 #SBATCH --time=00:05:00 #SBATCH --no-requeue #SBATCH --mail-type=FAIL srun hostname sleep 3
--cpus-per-task=1 | For SLURM, a CPU core (a CPU thread, to be more precise) is a CPU. |
--no-requeue | Prevent the job from being requeued after a failure. |
--mail-type=FAIL | Send an e-mail if sth. goes wrong. |
The srun command is responsible for the distribution of the program (hostname in our case) across the allocated resources, so that 80 instances of hostname will run on each of the allocated nodes concurrently. Please note, that this is not the only way to run or to distribute your processes. Other cases and methods are covered later in this document. In contrast, the sleep command is executed only on the head2) node.
Although nodes are allocated exclusively, you should always specify a memory value that reflects the RAM requirements of your job. The scheduler treats RAM as a consumable resource. As a consequence, if you omit the 3) Moreover, jobs are killed through SLURM's memory enforcement when using more memory than requested.
--nodes parameter (so that only the number of CPU cores is defined) and allocate more memory per core than there actually is on a node, you'll automatically get more nodes if the job doesn't fit in otherwise.
As already mentioned, after saving the above job script as e.g. jobscript.sh, you can submit your job by running
sbatch jobscript.sh
on the command line. The job's output streams (stdout and stderr) will be joined and saved to slurm-ID.out, where ID is a SLURM job ID, which is assigned automatically. You can change this behavior by adding an --output and/or --error argument to the SLURM options.
Job Monitoring
For job monitoring (to check the current state of your jobs) you can use the squeue command. Depending on the current cluster utilization (and other factors), your job(s) may take a while to start. You can list the current queuing times by running sqtimes on the command line.
If you need to cancel a job, you can use the scancel command (please see the manpage, man scancel, for further details).
Node Types
On Goethe-NHR four different types of compute nodes are available. There are
| Number | Type | Vendor | CPU | GPU | Cores per CPU | Cores per Node | Threads per Node | RAM [GB] |
|---|---|---|---|---|---|---|---|---|
| 412 | dual-socket | Intel | Xeon Skylake Gold 6148 | n/a | 20 | 40 | 80 | 192 |
| 72 | dual-socket | Intel | Xeon Skylake Gold 6148 | n/a | 20 | 40 | 80 | 772 |
| 139 | dual-socket | Intel | Xeon Broadwell E5-2640 v4 | n/a | 10 | 20 | 40 | 128 |
| 112 | dual-socket | AMD | EPYC 7452 | 8x MI210 | 32 | 64 | 128 | 512 |
In order to separate the node types, we employ the concept of partitions. We provide three partitions for the nodes, one for the Skylake CPU nodes, one for the Broadwell and one for the GPU nodes. Furthermore there is a small test partition (8 nodes). You have to select the node type you prefer by setting the partition:
| Partition | Partition/feature | CPU |
|---|---|---|
| general1 | #SBATCH --partition=general1 | Intel Skylake CPU |
| general2 | #SBATCH --partition=general2 #SBATCH --constraint=broadwell | Intel Broadwell CPU |
| gpu | #SBATCH --partition=gpu | AMD EPYC 7452 |
| test | #SBATCH --partition=test | Intel Skylake CPU |
Per-User Resource Limits
On Goethe-NHR, you have the following limits for the partitions general1 and general2:
| Limit | Value | Description |
|---|---|---|
MaxTime | 21 days | the maximum run time for jobs |
MaxJobsPU | 40 | max. number of jobs a user is able to run simultaneously |
MaxSubmitPU | 50 | max. number of jobs in running or pending state |
MaxNodesPU | 150 | max. number of nodes a user is able to use at the same time |
MaxArraySize | 1001 | the maximum job array size |
For the partition gpu we have following limits:
| Limit | Value | Description |
|---|---|---|
MaxTime | 21 days | the maximum run time for jobs |
MaxJobsPU | 20 | max. number of jobs a user is able to run simultaneously |
MaxSubmitPU | 30 | max. number of jobs in running or pending state |
MaxNodesPU | 20 | max. number of nodes a user is able to use at the same time |
For the partition test we have following limits:
| Limit | Value | Description |
|---|---|---|
MaxTime | 2 hours | the maximum run time for jobs |
MaxJobsPU | 2 | max. number of jobs a user is able to run simultaneously |
MaxSubmitPU | 3 | max. number of jobs in running or pending state |
MaxNodesPU | 2 | max. number of nodes a user is able to use at the same time |
GPU Jobs
Since December 2020, GPU accelerator nodes are part of the cluster (please see Hardware and Node Types). Select the gpu partition by setting
#SBATCH --partition=gpu
There is also a GPU test partition for compiling or testing your code. You can access it by setting e.g.
#SBATCH --partition=gpu_test #SBATCH --nodes=1 #SBATCH --ntasks=1 #SBATCH --gres=gpu:1 #SBATCH --mem=150g #SBATCH --time=01:00:00
in your allocation (see also: The salloc Command). In gpu_test, there are two GPU nodes. The max. time limit is 8 hours. Please also note, that nodes are shared here (in contrast to the gpu partition, where nodes are allocated exclusively), so that multiple jobs can run on the same node.
Hyper-Threading
On compute nodes you can use Hyper-Threading. That means, in addition to each physical CPU core a virtual core is available. SLURM identifies all physical and virtual cores of a node, so that you have 80 logical CPU cores on an Intel Skylake node, 40 logical CPU cores on an Intel Broadwell or Ivy Bridge node, and 128 logical CPU cores on a GPU node. If you don't want to use HT, you can disable it by adding
| Node type | hyperthreading=OFF | #cores per node | hyperthreading=ON | #cores per node |
|---|---|---|---|---|
| Skylake | #SBATCH --extra-node-info=2:20:1 | 40 | #SBATCH --extra-node-info=2:20:2 | 80 |
| Broadwell / Ivy Bridge | #SBATCH --extra-node-info=2:10:1 | 20 | #SBATCH --extra-node-info=2:10:2 | 40 |
| AMD EPYC 7452 | #SBATCH --extra-node-info=2:32:1 | 64 | #SBATCH --extra-node-info=2:32:2 | 128 |
to your job script. Then you'll get half the threads per node (which will correspond to the number of cores). This can be beneficial in some cases (some jobs may run faster and/or more stable).
Bundling Single-Threaded Tasks
Note: Please also see the Job Arrays section below. Because only full nodes are given to you, you have to ensure, that the available resources are used efficiently. Please combine as many single-threaded jobs as possible into one. The limits for the number of combined jobs are given by the number of cores and the available memory. A simple job script to start 40 independent processes may look like this one:
#!/bin/bash #SBATCH --partition=general1 #SBATCH --nodes=1 #SBATCH --ntasks=40 #SBATCH --cpus-per-task=1 #SBATCH --mem=128g #SBATCH --time=01:00:00 #SBATCH --mail-type=FAIL # # Replace by a for loop. ./program input01 &> 01.out & ./program input02 &> 02.out & ... ./program input40 &> 40.out & # Wait for all child processes to terminate. wait
In this (SIMD) example we assume, that there is a program (called program) which is run 40 times on 40 different inputs (usually input files). Both output streams (stdout and stderr) of each process are redirected to a file N.out. A job script is always executed on the first allocated node, so we don't need to use srun, since exactly one node is allocated. Further we assume that the executable is located in the same directory where the job was submitted (the initial working directory).
Job Arrays
If you have lots of single-core computations to run, job arrays are worth a look. Telling SLURM to run a job script as a job array will result in running that script multiple times (after the corresponding resources have been allocated). Each instance will have a distinct SLURM_ARRAY_TASK_ID variable defined in its environment.
Due to our full-node policy, you still have to ensure, that your jobs don't waste any resources. Let's say, you have 320 single-core tasks. In the following example 320 tasks are run inside a job array while ensuring that only 40-core nodes are used and that each node runs exactly 40 tasks in parallel.
#!/bin/bash #SBATCH --partition=general1 #SBATCH --nodes=1 #SBATCH --ntasks=40 #SBATCH --cpus-per-task=1 #SBATCH --mem-per-cpu=2000 #SBATCH --time=00:10:00 #SBATCH --array=0-319:40 #SBATCH --mail-type=FAIL my_task() { # Print the given "global task number" with leading zeroes # followed by the hostname of the executing node. K=$(printf "%03d" $1) echo "$K: $HOSTNAME" # Do nothing, just sleep for 3 seconds. sleep 3 } # # Every 40-task block will run on a separate node. for I in $(seq 40); do # This is the "global task number". Since we have an array of # 320 tasks, J will range from 1 to 320. J=$(($SLURM_ARRAY_TASK_ID+$I)) # Put each task into background, so that tasks are executed # concurrently. my_task $J & # Wait a little before starting the next one. sleep 1 done # Wait for all child processes to terminate. wait
If the task running times vary a lot, consider using the thread pool pattern. Have a look at GNU parallel, for instance.
OpenMP Jobs
For OpenMP jobs, set the --cpus-per-task parameter. You could specify a --mem-per-cpu value. But in this case you have to divide the total RAM required by your program by the number of threads. E.g. if your application needs 8000 MB and you want to run 40 threads, then you have to set --mem-per-cpu=200 (8000/40 = 200). However, it's also possible to specify the total amount of RAM using the --mem parameter. Don't forget to set the OMP_NUM_THREADS environment variable. Example:
#!/bin/bash #SBATCH --partition=general1 #SBATCH --nodes=1 #SBATCH --ntasks=1 #SBATCH --cpus-per-task=40 #SBATCH --mem=8000 #SBATCH --mail-type=ALL #SBATCH --time=48:00:00 export OMP_NUM_THREADS=$SLURM_CPUS_PER_TASK ./your_omp_program
MPI Jobs
Remember: Nodes are used exclusively. Each node has many CPU cores. If you want to run small jobs (i.e. where more than one job could be run on a single node concurrently), consider running more than one computation within a job. Otherwise it will most likely result in a waste of resources and will lead to a longer queueing time (for you and others).
See also: http://www.schedmd.com/slurmdocs/faq.html#steps
As an example, we want to run a program that spawns 80 MPI ranks and where 1200 MB of RAM are allocated for each rank.
#!/bin/bash #SBATCH --partition=general1 #SBATCH --nodes=2 #SBATCH --ntasks=80 #SBATCH --ntasks-per-node=40 #SBATCH --cpus-per-task=1 #SBATCH --mem-per-cpu=1200 #SBATCH --mail-type=ALL #SBATCH --extra-node-info=2:20:1 # Don't use this with Intel MPI. #SBATCH --time=48:00:00 module load mpi/.../<version> mpirun ./your_mpi_program
--mem-per-cpu is used for a job allocation and --mem-per-cpu times the number of CPUs on a node is greater than the total memory of that node. Please see Memory Allocation.
Some MPI installations support the srun command (instead of or in addition to mpirun), e.g.:
[...] module load mpi/.../<version> srun --mpi=pmix ./your_mpi_program
MPI implementations are typically designed to work seamlessly with job schedulers like Slurm. When you launch MPI tasks with mpirun (or srun) inside your job script, the MPI library uses the information provided by Slurm (via environment variables or other means) to determine the communication topology and allocate processes accordingly.
Hybrid Jobs: MPI/OpenMP
MPI example script (40 ranks, 6 threads each and 200 MB per thread, i.e. 1.2 GB per rank; so, for 40*6 threads, you'll get six 40-core nodes):
#!/bin/bash #SBATCH --partition=general1 #SBATCH --ntasks=40 #SBATCH --cpus-per-task=6 #SBATCH --mem-per-cpu=200 #SBATCH --mail-type=ALL #SBATCH --extra-node-info=2:20:1 # Don't use this with Intel MPI. #SBATCH --time=48:00:00 module load mpi/.../<version> export OMP_NUM_THREADS=6 # When using MVAPICH2 disable core affinity. export MV2_ENABLE_AFFINITY=0 mpirun -np 40 ./example_program
Please note, that this is just an example. You may or may not run it as-it-is with your software, which is likely to have a different scalability.
You have to disable the core affinity when running hybrid jobs with MVAPICH24). Otherwise all threads of an MPI rank will be pinned to the same core. Our example now includes the command
export MV2_ENABLE_AFFINITY=0
which disables this feature. The OS scheduler is now responsible for the placement of the threads during the runtime of the program. But the OS scheduler can dynamically change the thread placement during the runtime of the program. This leads to cache invalidation, which degrades performance. This can be prevented by thread pinning (topic not covered here).
When using Intel MPI, please also check its pinning parameters.
Memory Allocation
Normally the memory available per CPU thread is calculated by the whole amount of RAM divided by the number of threads. On GOETHE you can choose between four different types of nodes. For instance we select one Sylake node with 192GB of memory and divide it by 80 threads which is 2.4GB per thread 5). Keep in mind that the GOETHE clusters provides two threads per core. On GOETHE you have the convenience to select nodes with more memory but there is also another way to accomplish your job on an ordinary node with 192GB. Now imagine you need more memory as provided due to the selection of an ordinary Skylake node with 2.4GB per thread, we assume 8192MB per task. Type scontrol show node=<node> and look for 'mem', which is 187,5G(B). Another way is to login into a Skylake Node and type free -m, which states 192089M(B).6) Now we calculate how many processes we can launch on one node like this: 192089MB/8192MB=23.44… . With this result we can determine how many nodes we need. In the following example we like to have 71 processes à 8192MB.
#!/bin/bash
#SBATCH --job-name=<your_job_name>
#SBATCH --partition=general1
#SBATCH --ntasks=71 # Whole amount of processes we have.
#SBATCH --cpus-per-task=1 # Only one task per CPU.
# #SBATCH --mem-per-cpu=8192 # We can't use this argument here, because it ends up with an error
# message, therefore it's commented out.
#SBATCH --mem=0 # Use this argument instead, which means "full memory of the node".
#SBATCH --ntasks-per-node=23 # We have calculated and rounded that we need 23 per node.
srun hostname
If everything works fine you were granted 4 nodes. For example 3 nodes à 22 tasks and 1 node à 5 tasks, 66 tasks + 5 tasks = 71 tasks, as requested.
Please keep in mind, that you might waste resources because you could have used 80 threads but only use 23/80 due to your memory requirement.
Local Storage
On each node there is up to 1.4 TB of local disk space (see also Storage). If you need local storage, you have to add the --tmp parameter to your SLURM script. Set the amount of storage in megabytes, e.g. set --tmp=5000 to allocate 5 GB of local disk space. The data in the local directory (/local/$SLURM_JOB_ID) is automatically deleted after the corresponding batch job has finished.
The salloc Command
For interactive workflows you can use SLURM's salloc command. With salloc almost the same options can be used as with sbatch, e.g.:
[user@loginnode ~]$ salloc --nodes=4 --time=0:45:00 --mem=100g --partition=test salloc: Granted job allocation 197553 salloc: Waiting for resource configuration salloc: Nodes node45-[002-005] are ready for job [user@loginnode ~]$
Now you can ssh to the nodes that were allocated for the job and run further commands, e.g.:
[user@loginnode ~]$ ssh node45-002 [user@node45-002 ~]$ hostname node45-002.cm.cluster [user@node45-002 ~]$ logout Connection to node45-002 closed.
Or you can use srun for running a command on all allocated nodes in parallel:
[user@loginnode ~]$ srun hostname node45-002.cm.cluster node45-003.cm.cluster node45-005.cm.cluster node45-004.cm.cluster
Finally you can terminate your interactive job session by running exit, which will free the allocated nodes:
[user@loginnode ~]$ exit salloc: Relinquishing job allocation 197553
Planning Work
By using the --begin option it's possible to tell SLURM that you need the resources at some point in the future. Also, you might find it useful to use this feature for creating “user-mode reservations”. E.g.
- Submit a sleep job (allocate twenty intel20 nodes for 3 days), you can logout after running this command (but check the output of the squeue command first, if there is no corresponding pending job, then sth. went wrong):
$ sbatch --begin=202X-07-23T08:00 --time=3-0 --nodes=20 \ --partition=general2 --mem=120g \ --wrap="sleep 3d"
- Wait until the time has come (07/23/202X 8:00am or later, there is no guarantee, that the allocation will be made on time, but the earlier you submit the job, the more likely you'll get the resources by that time).
- Find out whether the sleep job is running (i.e. is in R state) and run a new job step within that allocation (see also http://slurm.schedmd.com/faq.html#multi_batch):
$ squeue JOBID PARTITION NAME ST TIME NODES 2717365 parallel sbatch R 3:28:29 20 $ srun --jobid 2717365 hostname ...
Note: Please note, that we are using thesruncommand. Thesbatchcommand is not supported in this scenario. - Finally, don't forget to release the allocation, if there's time left and the sleep job is still running:
$ scancel 2717365
Special software
Install R in ${HOME}
Howto install a local R from scratch in your /home directory without root access:
- Go to the main page https://cran.r-project.org. There you see the latest release. Copy the link.
- Go to to your preferred directory and do a
wget https://cran.r-project.org/src/base/R-<X>/R-<X.Y.Z>.tar.gz
tar xvf R-<X>/R-<X.Y.Z>.tar.gz
- Go to
R-<X.Y.Z> - In your browser address bar type https://cran.r-project.org/doc/FAQ/R-FAQ.html or https://cran.r-project.org/doc/FAQ/R-FAQ.html#How-can-R-be-installed-_0028Unix_002dlike_0029 to read some interesting things about R and Linux.
- Go to your terminal and type:
./configure --prefix=${HOME}/where/you/want/R/to/go make make install make installis not mandatory, you could go toR-<X.Y.Z>/binand callR, butmake installworks with your prefix directory.- Go to
${HOME}/prefix_R_directory/bin - Don't forget to add the R directory to your path:
export PATH=prefix_R_directory:${PATH} - Type
./R - Done.
R script for parallelization
Run with ''Rscript test.r''
# Rscript test.r library(foreach) library(doParallel) library(MASS) starts <- rep(100, 40) fx <- function(nstart) kmeans(Boston, 4, nstart=nstart) numCores <- detectCores() numCores system.time( results <- lapply(starts, fx) ) system.time( results <- mclapply(starts, fx, mc.cores = numCores) ) x <- iris[which(iris[,5] != "setosa"), c(1,5)] trials <- seq(1, 10000) boot_fx <- function(trial) { ind <- sample(100, 100, replace=TRUE) result1 <- glm(x[ind,2]~x[ind,1], family=binomial(logit)) r <- coefficients(result1) res <- rbind(data.frame(), r) } system.time({ results <- mclapply(trials, boot_fx, mc.cores = numCores) }) registerDoParallel(numCores) # use multicore, set to the number of our cores foreach (i=1:3) %dopar% { sqrt(i) } # Return a vector foreach (i=1:3, .combine=c) %dopar% { sqrt(i) } # Return a data frame foreach (i=1:3, .combine=rbind) %dopar% { sqrt(i) } # Let's use the iris data set to do a parallel bootstrap # From the doParallel vignette, but slightly modified x <- iris[which(iris[,5] != "setosa"), c(1,5)] trials <- 10000 system.time({ r <- foreach(icount(trials), .combine=rbind) %dopar% { ind <- sample(100, 100, replace=TRUE) result1 <- glm(x[ind,2]~x[ind,1], family=binomial(logit)) coefficients(result1) } }) # And compare that to what it takes to do the same analysis in serial system.time({ r <- foreach(icount(trials), .combine=rbind) %do% { ind <- sample(100, 100, replace=TRUE) result1 <- glm(x[ind,2]~x[ind,1], family=binomial(logit)) coefficients(result1) } }) stopImplicitCluster()